-
PDF
- Split View
-
Views
-
Cite
Cite
Andrea Kovacs-Simon, Rosanna Leuzzi, Magdalena Kasendra, Nigel Minton, Richard W. Titball, Stephen L. Michell, Lipoprotein CD0873 Is a Novel Adhesin of Clostridium difficile, The Journal of Infectious Diseases, Volume 210, Issue 2, 15 July 2014, Pages 274–284, https://doi.org/10.1093/infdis/jiu070
Close - Share Icon Share
ABSTRACT
Clostridium difficile is a cause of antibiotic-associated diarrhea and colitis, a healthcare-associated intestinal disease. Colonization of the gut is a critical step in the course of infection. The C. difficile lipoprotein CD0873 was identified as a putative adhesin through a bioinformatics approach. Surface exposure of CD0873 was confirmed and a CD0873 mutant was generated. The CD0873 mutant showed a significant reduction in adherence to Caco-2 cells and wild-type bacteria preincubated with anti-CD0873 antibodies showed significantly decreased adherence to Caco-2 cells. In addition, we demonstrated that purified recombinant CD0873 protein alone associates with Caco-2 cells. This is the first definitive identification of a C. difficile adhesin, which now allows work to devise improved measures for preventing and treating disease.
The gram-positive, anaerobic, spore-forming bacterium Clostridium difficile is a major cause of antibiotic-associated diarrhea and colitis and is a significant burden to healthcare services across Europe and North America [1, 2]. C. difficile infection (CDI) results in an estimated cost of $1.1 billion per year in the United States alone, as a consequence of patient treatment and time spent in hospital [3]. Gastrointestinal carriage of C. difficile may be asymptomatic or lead to disease, ranging from mild diarrhea to, at worst, potentially fatal pseudomembranous colitis [4]. CDI is primarily mediated by 2 large clostridial toxins (TcdA and TcdB), which cause fluid accumulation in the gut, inflammation, and destruction of the intestinal epithelium [5]. In addition, a binary toxin, CDT, is produced by several strains and has been suggested to induce morphological changes in intestinal epithelial cells, facilitating mucosal association of C. difficile [6].
Colonization is an early and critical step in the pathogenic process of C. difficile. In a hamster infection model, colonization by a nontoxigenic strain of C. difficile prevented animals from developing disease following a subsequent challenge with a toxigenic strain [7]. The interactions of bacterial surface proteins with intestinal tissues of the host may facilitate adherence of this pathogen to the gut. Therefore, surface proteins as colonization factors may contribute to the virulence of C. difficile [5]. Although a number of colonization factors have been identified and characterized in other bacterial pathogens, little is known about the colonization mechanisms of C. difficile. To date, several putative adhesins have been proposed [8–11], but as yet a genetic approach has not supported the role of these molecules in adherence.
Previous studies have shown that lipoprotein components of ABC-type transporters are frequently associated with the attachment of pathogenic bacteria, including group B Streptococcus [12], Brucella species [13], and Agrobacterium tumefaciens [14], to host cells. The multifunctional lipoprotein PsaA (protein surface antigen A) of Streptococcus pneumoniae is one of the best characterized ABC-transporter lipoprotein adhesins and is currently extensively studied as a potential vaccine component [15]. Apart from PsaA, PpmA, and SlrA are 2 other lipoproteins of S. pneumoniae that have been shown to contribute to virulence by promoting pneumococcal colonization [16, 17]. Furthermore, the extracellular matrix binding proteins DbpA (decorin) and BmpA (laminin), as well as OmpA, are all lipoproteins of Borrelia burgdorferi that have a contributory role in adherence [18–20]. Similarly, the adhesin BopA of Bifidobacterium bifidum [21] is a lipoprotein, as is the well-characterized adhesin IlpA of Vibrio vulnificus [22]. Numerous studies have demonstrated the contribution of lipoproteins to adherence of various bacterial pathogens, highlighting their importance in virulence, and these have been reviewed elsewhere [23].
To date, no study has investigated the role of the lipoproteins of C. difficile in adherence. In this work, we investigated whether a novel lipoprotein of C. difficile, CD0873, is involved in adhesion to host cells. We demonstrate that CD0873 is exposed at the bacterial cell surface and that the adherence of C. difficile to host cells is affected by mutation of the gene encoding CD0873. Moreover, we used antibodies against CD0873 to demonstrate the specificity of this adherence, providing additional support to the role of CD0873 as an adhesin of C. difficile.
MATERIALS AND METHODS
Bioinformatic Analysis
Lipoproteins of C. difficile 630 (RefSeq: NC_009089) were identified by the following criteria: (1) all phosphatidate cytidylyltransferase signatures were matched against the PROSITE pattern PS00013/PDOC00013, implemented as the PERL regular expression: [ACFGHILMNOPQSTUVXYZ]{6}[LIVMFWSTAG]{2}[LIVMFYSTAGCQ][AGS]C/[24]; (2) presence of a cysteine residue between the 15th and 35th amino acids; and (3) presence of at least 1 lysine or 1 arginine in the first 7 residues. The presence of a signal peptide was checked using SignalP 3.0 [25]. Sequence alignment of amino acid sequences was performed with CloneManager, using BLOSUM62 as a scoring matrix.
Bacterial Strains and Growth Conditions
C. difficile strains were grown at 37°C in an anaerobic workstation (Don Whitley, United Kingdom) in an atmosphere of 10% CO2, 10% H2, and 80% N2, using prereduced brain heart infusion broth or agar (Oxoid). Escherichia coli strains were routinely cultured in Luria-Bertani (LB) broth or on LB agar. When appropriate, antibiotics were added at the following concentrations: kanamycin, 50 µg/mL; tetracycline, 10 µg/mL; chloramphenicol, 12.5 µg/mL (broth) and 25 µg/mL (agar); cycloserine, 250 µg/mL; cefoxitin, 8 µg/mL; thiamphenicol, 15 µg/mL; and erythromycin, 5 µg/mL. Strains, plasmids, and primers used in this study are described in Table 1.
Strains, Plasmids, and Primers Used in This Study
| Strains . | Description . | Reference . |
|---|---|---|
| C. difficile | ||
| 630Δerm | C. difficile wild-type strain | [26] |
| KSA1 | C. difficile CD0873::ermB::317¦318; Eryr | This study |
| KSA2 | C. difficile CD0873::ermB::317¦318, carrying pMTL960-CD0873; Eryr, Thir | This study |
| fliC mutant | C. difficile fliC::ermB::515¦516; Eryr | [27] |
| E. coli | ||
| DH5α | Cloning host | Invitrogen |
| C43(DE3) | Expression host | [28] |
| CA434 | Conjugation donor; Kanr, Tetr | [29] |
| Plasmids | ||
| pNIC28-Bsa4 | His-tag protein expression vector; Kanr | [30] |
| pNIC-KSA1 | pNIC28 carrying CD0873 | This study |
| pMTL007C-E2 | Shuttle vector; Chlr, Thir | [31] |
| pMTL-KSA1 | pMTL007C-E2 carrying the targeted intron for CD0873 at 317¦318 site | This study |
| pRPF144 | pMTL960-based shuttle plasmid vector carrying the promoter of the cwp gene and the gusA gene, Chlr, Thir | [32] |
| pMTL-KSA2 | pRPF144 carrying the CD0873 gene (instead of gusA) | This study |
| Primers (5′ to >3′) | ||
| pNIC_CD0873-F | TACTTCCAATCCATGGCTTCACAAGGAGGAGACTCTGG | |
| pNIC_CD0873-R | TATCCACCTTTACTGTCATTCTTGTTTAGTCTTTACAT | |
| CD0873-F | CAGCATTAGATGCCACTAGG | |
| CD0873-R | GCAGCAACAGGGTCACTCAC | |
| Compl_CD0873-F | GAGCTCTTTTTGAGGGGGATATAAAAATG | |
| Compl_CD0873-R | GGATCCCTATTCTTGTTTAGTCTTTAC | |
| RT-PCR_maa-F | TGTTGTCTGGGAAAGGATAC | |
| RT-PCR_maa-R | TGGATTTCCAACAGCTACAG | |
| RT-PCR_CD0873-F | GCTGGTGCTCTTAGTATATC | |
| RT-PCR_CD0873-R | GATGTAGCCACAGGCATATC | |
| RT-PCR_CD0874-F | GGAACTTGCCCTTCTATGAC | |
| RT-PCR_CD0874-R | GGGTCAAGTGCTGCTGTATG | |
| Strains . | Description . | Reference . |
|---|---|---|
| C. difficile | ||
| 630Δerm | C. difficile wild-type strain | [26] |
| KSA1 | C. difficile CD0873::ermB::317¦318; Eryr | This study |
| KSA2 | C. difficile CD0873::ermB::317¦318, carrying pMTL960-CD0873; Eryr, Thir | This study |
| fliC mutant | C. difficile fliC::ermB::515¦516; Eryr | [27] |
| E. coli | ||
| DH5α | Cloning host | Invitrogen |
| C43(DE3) | Expression host | [28] |
| CA434 | Conjugation donor; Kanr, Tetr | [29] |
| Plasmids | ||
| pNIC28-Bsa4 | His-tag protein expression vector; Kanr | [30] |
| pNIC-KSA1 | pNIC28 carrying CD0873 | This study |
| pMTL007C-E2 | Shuttle vector; Chlr, Thir | [31] |
| pMTL-KSA1 | pMTL007C-E2 carrying the targeted intron for CD0873 at 317¦318 site | This study |
| pRPF144 | pMTL960-based shuttle plasmid vector carrying the promoter of the cwp gene and the gusA gene, Chlr, Thir | [32] |
| pMTL-KSA2 | pRPF144 carrying the CD0873 gene (instead of gusA) | This study |
| Primers (5′ to >3′) | ||
| pNIC_CD0873-F | TACTTCCAATCCATGGCTTCACAAGGAGGAGACTCTGG | |
| pNIC_CD0873-R | TATCCACCTTTACTGTCATTCTTGTTTAGTCTTTACAT | |
| CD0873-F | CAGCATTAGATGCCACTAGG | |
| CD0873-R | GCAGCAACAGGGTCACTCAC | |
| Compl_CD0873-F | GAGCTCTTTTTGAGGGGGATATAAAAATG | |
| Compl_CD0873-R | GGATCCCTATTCTTGTTTAGTCTTTAC | |
| RT-PCR_maa-F | TGTTGTCTGGGAAAGGATAC | |
| RT-PCR_maa-R | TGGATTTCCAACAGCTACAG | |
| RT-PCR_CD0873-F | GCTGGTGCTCTTAGTATATC | |
| RT-PCR_CD0873-R | GATGTAGCCACAGGCATATC | |
| RT-PCR_CD0874-F | GGAACTTGCCCTTCTATGAC | |
| RT-PCR_CD0874-R | GGGTCAAGTGCTGCTGTATG | |
Abbreviations: C. difficile, Clostridium difficile; E. coli, Escherichia coli.
Strains, Plasmids, and Primers Used in This Study
| Strains . | Description . | Reference . |
|---|---|---|
| C. difficile | ||
| 630Δerm | C. difficile wild-type strain | [26] |
| KSA1 | C. difficile CD0873::ermB::317¦318; Eryr | This study |
| KSA2 | C. difficile CD0873::ermB::317¦318, carrying pMTL960-CD0873; Eryr, Thir | This study |
| fliC mutant | C. difficile fliC::ermB::515¦516; Eryr | [27] |
| E. coli | ||
| DH5α | Cloning host | Invitrogen |
| C43(DE3) | Expression host | [28] |
| CA434 | Conjugation donor; Kanr, Tetr | [29] |
| Plasmids | ||
| pNIC28-Bsa4 | His-tag protein expression vector; Kanr | [30] |
| pNIC-KSA1 | pNIC28 carrying CD0873 | This study |
| pMTL007C-E2 | Shuttle vector; Chlr, Thir | [31] |
| pMTL-KSA1 | pMTL007C-E2 carrying the targeted intron for CD0873 at 317¦318 site | This study |
| pRPF144 | pMTL960-based shuttle plasmid vector carrying the promoter of the cwp gene and the gusA gene, Chlr, Thir | [32] |
| pMTL-KSA2 | pRPF144 carrying the CD0873 gene (instead of gusA) | This study |
| Primers (5′ to >3′) | ||
| pNIC_CD0873-F | TACTTCCAATCCATGGCTTCACAAGGAGGAGACTCTGG | |
| pNIC_CD0873-R | TATCCACCTTTACTGTCATTCTTGTTTAGTCTTTACAT | |
| CD0873-F | CAGCATTAGATGCCACTAGG | |
| CD0873-R | GCAGCAACAGGGTCACTCAC | |
| Compl_CD0873-F | GAGCTCTTTTTGAGGGGGATATAAAAATG | |
| Compl_CD0873-R | GGATCCCTATTCTTGTTTAGTCTTTAC | |
| RT-PCR_maa-F | TGTTGTCTGGGAAAGGATAC | |
| RT-PCR_maa-R | TGGATTTCCAACAGCTACAG | |
| RT-PCR_CD0873-F | GCTGGTGCTCTTAGTATATC | |
| RT-PCR_CD0873-R | GATGTAGCCACAGGCATATC | |
| RT-PCR_CD0874-F | GGAACTTGCCCTTCTATGAC | |
| RT-PCR_CD0874-R | GGGTCAAGTGCTGCTGTATG | |
| Strains . | Description . | Reference . |
|---|---|---|
| C. difficile | ||
| 630Δerm | C. difficile wild-type strain | [26] |
| KSA1 | C. difficile CD0873::ermB::317¦318; Eryr | This study |
| KSA2 | C. difficile CD0873::ermB::317¦318, carrying pMTL960-CD0873; Eryr, Thir | This study |
| fliC mutant | C. difficile fliC::ermB::515¦516; Eryr | [27] |
| E. coli | ||
| DH5α | Cloning host | Invitrogen |
| C43(DE3) | Expression host | [28] |
| CA434 | Conjugation donor; Kanr, Tetr | [29] |
| Plasmids | ||
| pNIC28-Bsa4 | His-tag protein expression vector; Kanr | [30] |
| pNIC-KSA1 | pNIC28 carrying CD0873 | This study |
| pMTL007C-E2 | Shuttle vector; Chlr, Thir | [31] |
| pMTL-KSA1 | pMTL007C-E2 carrying the targeted intron for CD0873 at 317¦318 site | This study |
| pRPF144 | pMTL960-based shuttle plasmid vector carrying the promoter of the cwp gene and the gusA gene, Chlr, Thir | [32] |
| pMTL-KSA2 | pRPF144 carrying the CD0873 gene (instead of gusA) | This study |
| Primers (5′ to >3′) | ||
| pNIC_CD0873-F | TACTTCCAATCCATGGCTTCACAAGGAGGAGACTCTGG | |
| pNIC_CD0873-R | TATCCACCTTTACTGTCATTCTTGTTTAGTCTTTACAT | |
| CD0873-F | CAGCATTAGATGCCACTAGG | |
| CD0873-R | GCAGCAACAGGGTCACTCAC | |
| Compl_CD0873-F | GAGCTCTTTTTGAGGGGGATATAAAAATG | |
| Compl_CD0873-R | GGATCCCTATTCTTGTTTAGTCTTTAC | |
| RT-PCR_maa-F | TGTTGTCTGGGAAAGGATAC | |
| RT-PCR_maa-R | TGGATTTCCAACAGCTACAG | |
| RT-PCR_CD0873-F | GCTGGTGCTCTTAGTATATC | |
| RT-PCR_CD0873-R | GATGTAGCCACAGGCATATC | |
| RT-PCR_CD0874-F | GGAACTTGCCCTTCTATGAC | |
| RT-PCR_CD0874-R | GGGTCAAGTGCTGCTGTATG | |
Abbreviations: C. difficile, Clostridium difficile; E. coli, Escherichia coli.
Purification of Recombinant CD0873 Protein
Expression clones were generated by polymerase chain reaction (PCR), using primers pNIC_CD0873-F and pNIC_CD0873-R (Table 1) and ligation-independent cloning into vector pNIC28-Bsa4 [30]. The resultant pNIC-KSA1 plasmid was transformed into Escherichia coli C43(DE3) [28]. The histidine-tagged CD0873 was overexpressed by IPTG induction (0.5 mM), purified with a His-Bind kit (Novagen), and cleaved by TEV protease (Promega).
Production of CD0873-Specific Antibodies
All antisera were generated commercially (Enzo Life Sciences). Rabbits received a 50-µg dose of CD0873 7 times at 2-week intervals.
Protein Analysis
Whole-cell lysates of C. difficile were prepared as described previously [32]. Subsequent extraction with Triton X-114 enriched for hydrophobic proteins [33]. Protein samples (whole-cell lysates and aqueous- and detergent-phase proteins) were Western blotted and probed with CD0873-specific primary antibody (dilution, 1:2500) and anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (dilution, 1:2000) secondary antibody (Cell Signaling Technology). Blots were developed using LumiGLO chemiluminescent substrate (Cell Signaling Technology) and visualized using the Bio-Rad ChemiDoc system (Bio-Rad Laboratories).
Immunofluorescence Microscopy
Bacteria were washed with phosphate-buffered saline (PBS), fixed in 3.7% (w/v) paraformaldehyde (Sigma), and immobilized on poly-D-lysine slides. Slides were blocked with 3% (w/v) bovine serum albumin (BSA; Sigma) in PBS for 1 hour, incubated with CD0873-specific primary antibody (1:500) for 1 hour, and washed with PBS. Slides were then incubated with Alexa Fluor 568–labeled goat anti-rabbit IgG (dilution, 1:1000; Molecular Probes). Labeled samples were mounted with ProLong Gold antifade reagent with DAPI (Molecular Probes) and analyzed with a Zeiss LSM710 confocal microscope.
Generation and Complementation of a CD0873 Mutant of C. Difficile
A CD0873 mutant of C. difficile 630Δerm (KSA1) was produced using the ClosTron system [34]. Vector pMTL-KSA1, retargeted for the gene encoding CD0873, was purchased from DNA2.0. Putative mutants were screened using CD0873 gene–specific primers (CD0873-F and CD0873-R; Table 1) and verified by sequencing the amplicons. For complementation studies, the CD0873 gene and 20-bp of the 5′ region encompassing the Shino-Dalgarno sequence were amplified from genomic DNA, using Compl_CD0873-F and Compl_CD0873-R primers (Table 1). The PCR product was cloned into a pMTL960-based shuttle plasmid (pRPF144) containing the native promoter of the cwp2 gene, resulting in the plasmid pMTL-KSA2 [32]. pMTL-KSA2 was transformed into E. coli CA434 and conjugated into the KSA1 to generate KSA2.
Adherence Assay
Caco-2 cells were used between passages 54 and 69 and cultured in Eagle's minimal essential medium (EMEM; PromoCell, Heidelberg, Germany) supplemented with 10% fetal bovine serum (SAFC, Sigma-Aldrich) and 1% MEM nonessential amino acids (NEAA; Life Technologies) in a humidified 5% CO2 atmosphere at 37°C. Cells were seeded at 7 × 104 cells per well in 24-well tissue culture plates or on collagen I–coated chamber slides (BD Biocat) for immunofluorescence microscopy. Monolayers were used 8 days after seeding. The culture medium was replaced with nonsupplemented EMEM 24 hours before infection. Bacterial suspensions for infection of Caco-2 cells were prepared in prereduced EMEM, and adherence experiments were conducted under anaerobic conditions. Caco-2 monolayers were infected at a multiplicity of infection of 5 or 100. After 2 hours of incubation, nonadherent bacteria were removed by pipetting, and cells were washed 3 times with PBS. For the adherence-blocking experiments, bacterial cells were preincubated with CD0873 antisera for 30 minutes at various dilutions. Adherence of C. difficile was determined by 3 methods. First, colony-forming units (CFUs) were counted, wherein washed cells were resuspended in 1 mL of brain heart infusion broth, and the counts of adherent CFU were determined by plating. Second, an on-cell Western assay was performed. Here, Caco-2 cells with the adhered bacteria were fixed with 5% paraformaldehyde (Fisher Scientific) for 25 minutes at room temperature. Cells were washed 5 times with PBS and blocked overnight at 4°C with Odyssey blocking buffer (LI-COR). Cells were then incubated with anti-C. difficile antibodies (dilution, 1:2500; antibody was raised against whole cells of C. difficile 630, kindly provided by Prof N. Fairweather) for 1 hour, followed by incubation with IRdye 680LT goat anti-rabbit IgG (LI-COR; dilution, 1:2000) for 1 hour in the dark. Wells were washed with PBS, and fluorescence was measured by the Odyssey CLx infrared scanner (LI-COR). Third, immunofluorescence microscopy was conducted. In this method, cells were fixed with 3.7% paraformaldehyde (Sigma) for 15 minutes, washed, and blocked with 3% BSA in PBS for 1 hour. Slides were then incubated with anti-C. difficile antibodies (dilution, 1:1000) for 1 hour. Cells were then washed and incubated with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 568-phalloidin (dilutions, 1:1000 and 1:400, respectively) (Molecular Probes) for 1 hour. After successive washes with PBS, slides were mounted with ProLong Gold antifade reagent with DAPI (Molecular Probes) and analyzed with a Zeiss LSM710 confocal microscope.
Protein Binding Assays
These experiments were performed under aerobic conditions. After removal of EMEM and washing with PBS, Caco-2 cells were incubated with 1 mL of EMEM containing 100 µg of recombinant protein. After 3 washes with PBS, cells were disrupted with 200 µL of lysis buffer (0.8% sodium dodecyl sulfate in PBS) and boiled for 10 minutes. A 15-µL volume of the lysate was then subjected to Western blot analysis using anti-CD0873 and anti-His antibodies.
RESULTS
Identification of the CD0873 Lipoprotein
We first analyzed the genome of C. difficile strain 630 and identified 86 genes predicted to encode lipoproteins. We compared this list with the reported data on genes that are upregulated during infection of Caco-2 cells [35] and identified CD0873. The CD0873 gene product is predicted to be 340 amino acids and to contain a 23–amino acid lipoprotein signal peptide cleavage motif. The predicted CD0873 product showed 21% sequence identity to S. pneumoniae PsaA.
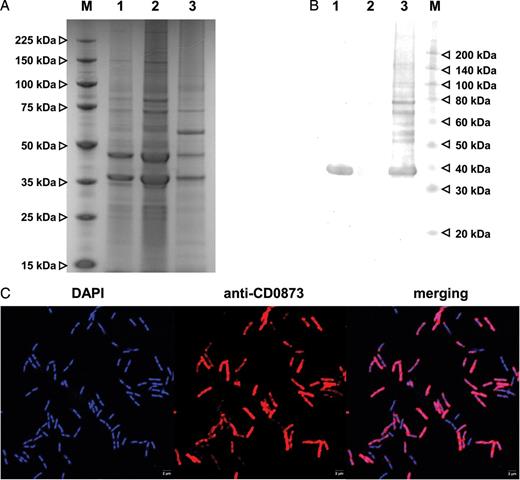
CD0873 Is a Lipoprotein Exposed on the Cell Surface of C. difficile
The CD0873 lipoprotein is exposed to the surface of Clostridium difficile. A, Sodium dodecyl sulfate–polyacrylamide gel electrophoresis of the whole-cell lysate and aqueous- and detergent-phase proteins of the wild-type C. difficile 630Δerm cells. B, Western blot analysis of the whole-cell lysate and aqueous- and detergent-phase proteins of the wild-type C. difficile 630Δerm cells, using anti-CD0873 antibodies. The CD0873 lipoprotein was detected in the detergent phase but not in the aqueous phase of the C. difficile 630Δerm cell lysate. Lane M, protein molecular weight marker (in kDa); lane 1, whole-cell lysate; lane 2, aqueous phase; lane 3, detergent phase. C, Immunofluorescence microscopy of the C. difficile 630Δerm strain, using anti-CD0873 antibodies and Fluo568-conjugated anti-rabbit secondary antibodies (red). DNA was stained with DAPI (blue). The CD0873 antibodies were able to bind to whole C. difficile 630Δerm wild-type cells.
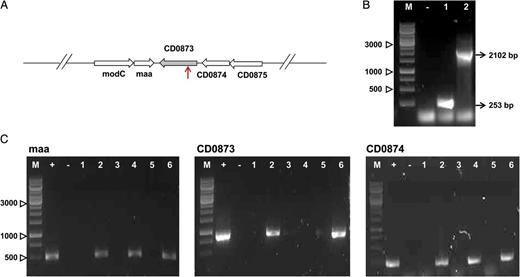
Construction of a CD0873 Mutant Strain
Construction and confirmation of the CD0873 mutant of Clostridium difficile. A, Schematic representation of the CD0873 region on the chromosome of C. difficile. The red arrow indicates the insertion site of the group II intron (1749 bp) into the CD0873 gene in the KSA1 strain (between the 317th and 318th bps). B, Confirmation of the CD0873 mutant by polymerase chain reaction (PCR), using CD0873-F and CD0873-R primers. Lane M, DNA ladder (in bp); lane 1, 630Δerm (253 bp); lane 2, KSA1 (2102 bp); −, water (negative) control. Sizes of the PCR products are indicated in brackets. C, Transcriptional analysis of the CD0873 gene and the genes upstream (CD0874) and downstream (maa) of the CD0873 gene in the 630Δerm, KSA1, and KSA2 strains by reverse-transcription PCR (RT-PCR), using RT-PCR primer pairs (Table 1). Lane M, DNA ladder (in bp); +, 630Δerm DNA (positive) control; −, water (negative) control; lanes 1, 3, and 5, DNAse treated RNA; lanes 2, 4, and 6, complementary DNA; lanes 1 and 2, 630Δerm; lanes 3 and 4, KSA1; lanes 5 and 6, KSA2. Sizes of the RT-PCR products are as follows: maa-RT F, R: 509 bp; CD0873-RT F, R: 884 bp; and CD0874-RT F, R: 255 bp. This analysis demonstrated that the mutation present in KSA1 did not affect the transcription of the CD0874 and the maa genes.
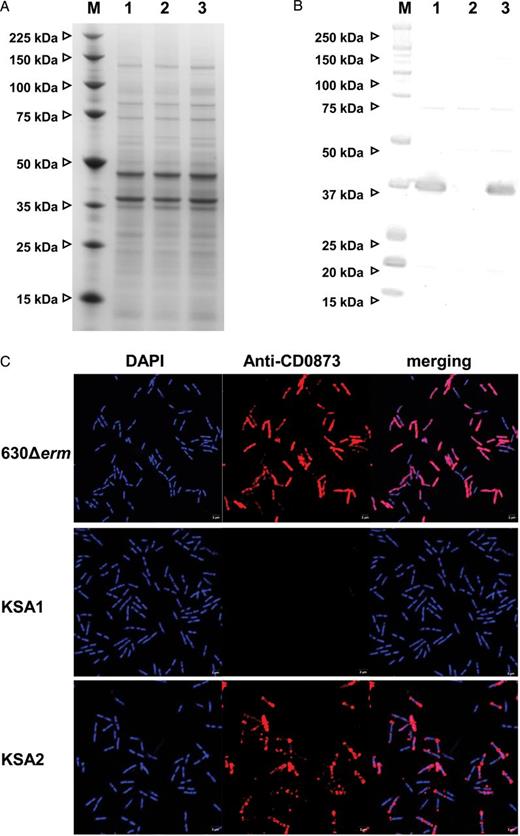
The CD0873 lipoprotein is not expressed in the CD0873 mutant of Clostridium difficile. A, Sodium dodecyl sulfate–polyacrylamide gel electrophoresis of the whole-cell lysates of the 630Δerm, KSA1, and KSA2 strains. B, Western blot analysis of the whole-cell lysates of the 630Δerm, KSA1, and KSA2 strains, using anti-CD0873 antibodies. The CD0873 lipoprotein was detected in 630Δerm and KSA2 but not in KSA1. Lane M, protein molecular weight marker (in kDa); lane 1, 630Δerm; lane 2, KSA1; lane 3, KSA2. C, Immunofluorescence microscopy of the 630Δerm, KSA1, and KSA2 cells, using anti-CD0873 antibodies and Fluo568-conjugated anti-rabbit secondary antibodies (red). DNA was stained with DAPI (blue). The CD0873 antibodies were able to bind to whole C. difficile 630Δerm wild-type and KSA2 cells but not to KSA1 cells.
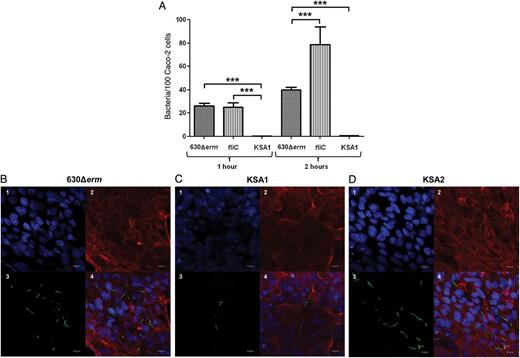
Mutation of CD0873 Abolishes Adherence of C. difficile to Caco-2 Cells
Inactivation of the CD0873 gene causes reduced adherence of Clostridium difficile to Caco-2 cells. A, Adherence of the C. difficile 630Δerm, fliC mutant, and KSA1 to Caco-2 monolayers, measured by colony-forming unit counts. Caco-2 cells were infected with the C. difficile strains (multiplicity of infection, 1:5) and incubated for 1 and 2 hours. The values are mean numbers of the adherent bacteria (with standard errors of the mean) per 100 Caco-2 cells of 4 separate experiments performed in triplicate. Statistical comparison of the value for 630Δerm to the value for fliC and KSA1 was performed by 1-way analysis of variance. ***P < .0001. B, Adherence of C. difficile 630Δerm, KSA1, and KSA2, assessed by confocal immunofluorescence microscopy. Caco-2 cells were incubated with the C. difficile strains (multiplicity of infection, 1:100) for 2 hours. Adherent bacteria were labeled by anti-C. difficile primary antibodies and secondary fluorescent antibodies (Alexa488, green). DNA and cellular actin were stained with DAPI (blue) and phalloidin-Alexa568 (red), respectively. The scale bar indicates 10 µm.
The role of CD0873 in adherence was thus further investigated by visualizing the adherence of C. difficile strains to Caco-2 cells by immunofluorescence microscopy. Compared with the wild-type strain, the adherence of C. difficile KSA1 was reduced (Figure 4B). In this assay, adherence was restored to wild-type levels in KSA2 (Figure 4C).
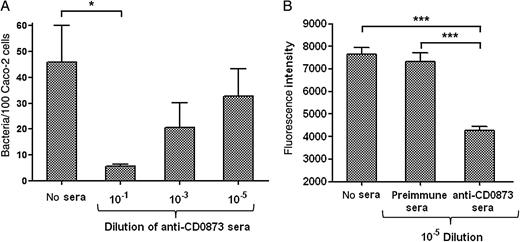
Cytoadherence of C. difficile Is Inhibited by Anti-CD0873 Antibodies
Adherence of Clostridium difficile to Caco-2 cells is inhibited by CD0873 antibodies. A, Adherence of wild-type C. difficile to Caco-2 monolayers after the bacterial cells were preincubated with CD0873 antisera. Cell-binding was measured by colony-forming unit counts. The values are mean numbers of the adherent bacteria per 100 Caco-2 cells. The assay was performed in duplicate in 3 independent experiments. The presented values are mean numbers (with standard errors of the mean). *P = .0132, compared with no-sera control, by the 2-tailed t test. B, Adherence of wild-type C. difficile to Caco-2 monolayers after preincubation of the bacteria with either CD0873 antisera or preimmune sera. Cell binding was determined by on-cell Western assay by measuring the fluorescence intensity, as described in Materials and Methods. Intensities indicated on the graph are normalized to the intensities measured with the Caco-2 cells–only control. ***P < .001, by 1-way analysis of variance (n = 5).
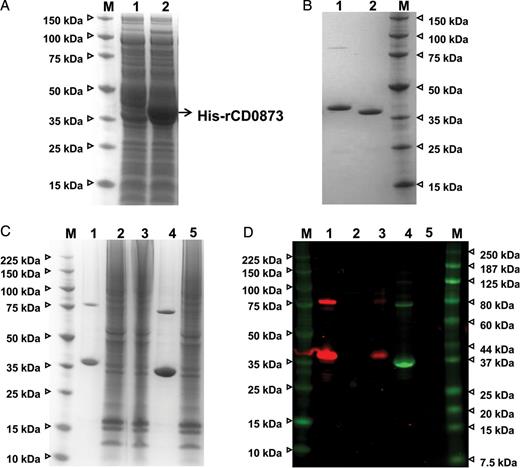
CD0873 Binds Directly to Caco-2 Cells
The recombinant CD0873 protein binds to Caco-2 cells. A, Expression of the recombinant CD0873 protein in Escherichia coli C43(DE3) by IPTG induction. Proteins of the Escherichia coli C43(DE3) cell lysates (lane 1, uninduced control; lane 2, induction of the protein expression with IPTG) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and stained with SimplyBlue SafeStain. M, protein molecular weight marker (in kDa). B, SDS-PAGE of the purified recombinant CD0873 protein. Following induction of protein expression with IPTG, the histidine-tagged recombinant protein was purified (lane 1), and the histidine tag was removed by treating the purified protein with TEV protease (lane 2). C, Loading control of the protein binding assay. Caco-2 cells (7 × 105) were incubated with 100 µg of recombinant CD0873 protein for 2 hours (no protein and recombinant ɛ prototoxin of Clostridium perfringens served as negative controls), the cells were then washed, and the proteins of the Caco-2 cell lysates that were analyzed for protein binding were separated by SDS-PAGE and stained with SimplyBlue SafeStain. D, CD0873 immunoblot of Caco-2 cell lysates by Western blotting, using rabbit anti-CD0873 antibodies (dilution, 1:2500). The control ɛ prototoxin was detected by mouse anti-His antibodies. The Caco-2 cells were bound by the recombinant CD0873 protein but not by the control protein. Lane 1, recombinant CD0873 protein; lane 2, lysate of Caco-2 cells plus no protein (negative control); lane 3, lysate of Caco-2 cells plus recombinant CD0873 protein; lane 4, recombinant ɛ prototoxin of C. perfringens; lane 5, lysate of Caco-2 cells plus recombinant ɛ prototoxin of C. perfringens (washing control); M, protein molecular weight marker (in kDa).
DISCUSSION
It is widely reported that colonization is crucial in C. difficile infection. Previous studies suggested that adherence of C. difficile to the gastrointestinal tract is multifactorial and involves several bacterial proteins, with other macromolecules possibly playing direct or indirect roles in attachment [5]. So far, only a few candidate proteins have been identified with probable or proven adhesin functions, binding either to extracellular matrix components or to gastrointestinal cells. However, when isogenic mutants have been constructed, they have failed to reveal a role for the putative adhesin in adherence to host cells [11, 36].
Molecules of C. difficile currently proposed to be involved in adhesion include surface-layer proteins, such as P36 and P47 [37, 38]; flagellar proteins [10]; cell wall proteins (cwp66 [39] and cwp84 [40]); Fbp68, a fibronectin binding protein [8]; heat shock protein GroEL [9]; and, more recently, a collagen binding protein [11]. Interestingly, absence of flagellar components (FliC and FliD) increases the in vitro adherence of C. difficile to intestinal Caco-2 cells. In contrast, it has been shown that fliC and fliD knockout mutants colonize the cecum at the same rate as the wild-type strain in a hamster infection model [27]. Thus, the role of flagella in adherence is currently unclear. Although it has been shown that C. difficile possesses a functional collagen binding protein, CbpA, mutation of cbpA failed to result in a reduction in adherence to IMR-90 cells, indicating that other factors of C. difficile may be involved in adhesion [11].
In other bacterial pathogens, lipoproteins have been shown to be essential adhesins and colonization factors [20, 41], with several of these classified as solute binding lipoproteins of ABC-type transport systems. Bioinformatic analysis led us to study the CD0873 protein, a polypeptide containing a lipoprotein signal peptide cleavage motif at its N-terminus and annotated as a sugar-family extracellular solute-binding lipoprotein of an ABC transport system. Partitioning of CD0873 to the detergent phase following a Triton X-114 phase extraction supports the annotation of this protein as a lipoprotein. Our immunofluorescence microscopy data reveal that CD0873 is surface exposed, probably anchored to the cell membrane through the attached acyl moieties. It has been proposed that the ability of lipoproteins to be N-terminus anchored to a lipid bilayer and simultaneously exposed on the surface of a bacterium is due to the presence of an extended N-terminus region, which, in gram-negative bacteria, can span the lipooligosaccharide layer [42].
The operon CD0875-CD0873, encoding for an ABC-type transporter, is highly conserved and present in all C. difficile strains for which genome sequence data are available. This operon shares the same genetic arrangement as the psaBCA operon that encodes for the streptococcal lipoprotein PsaA. This multifunctional protein is a major adhesin of S. pneumoniae, as well as a manganese ion transporter [43]. As to the function of CD0873, the protein has been annotated as an ABC transporter sugar-family extracellular solute-binding lipoprotein. Elucidation of the structure of CD0873 will aid in defining the role of this lipoprotein's function because of the possibility that CD0873 may share some functional similarity to the lipoprotein PsaA, owing to the conservation of some amino acids involved in manganese binding. Interestingly, expression of CD0874 was upregulated during infection of Caco-2 cells, suggesting that this operon maybe important in the interaction with enterocytes [35].
This latter finding directed us to generate a C. difficile mutant defective in CD0873 expression. On confirmation of this phenotype, we demonstrated that the mutant was unable to adhere to Caco-2 cells, suggesting that this protein may, similar to PsaA, function as an adhesin. Caco-2 cells are human colonic adenocarcinoma cells that differentiate spontaneously under standard culture conditions and resemble many in vivo characteristics of enterocytes [44]. Therefore, this cell line is commonly used as a validated human intestinal cell model for in vitro studies of host-pathogen interactions [41, 45], including those involving C. difficile and humans [46, 47]
More importantly, this finding is the first demonstration of a genetic deletion resulting in a clear reduction of C. difficile adherence in vitro. Furthermore, we could restore adherence by complementation, as evidenced by immunofluorescence microscopy (complementation in the as CFU assay was irreproducible, perhaps because of the different nature of this assay). This observation did not preclude the possibility that the lack of CD0873 may have an indirect effect on adhesion. To address this, we demonstrated that antibodies against the surface-exposed CD0873 could inhibit adherence. To further demonstrate that CD0873 is a bona fide adhesin, we used recombinant CD0873 in a direct binding assay to show that the protein has an intrinsic ability to interact with Caco-2 cells. Recently, it has been proposed that spores adhere to Caco-2 cells [48]. Given that CD0873 has been shown to be localized to the surface of spores [49], it raises the possibility that CD0873 might also play an important role during the early stages of colonization.
Given the lack of an effective toxin-based vaccine for C. difficile, the identification of novel factors key to the pathogenesis of this disease is essential. CD0873 has previously been identified as immunogenic in 3 of 6 patients infected with C. difficile, although no data relating to the severity of their infection were reported [50]. It would be of interest to look at a larger panel of patients with well-documented clinical findings to determine whether a high serum antibody response to CD0873 correlates with protection. Nevertheless, the data presented here support the hypothesis that CD0873 is a lipoprotein of C. difficile with a role in adherence. The finding that antibodies against CD0873 can inhibit adhesion raises the possibility that this protein could be a target for therapeutic intervention.
Notes
Financial support. This work was supported by the European Union Seventh Framework Programme (EU MCN Grant agreement 237942).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
Author notes
Presented in part: 4th International Clostridium difficile Symposium, Bled, Slovenia, 20–22 September 2012.